
Welcome!
PhyloCloud is an online platform for the interactive analysis, annotation and visualization of large collections of phylogenetic trees and multiple sequence alignments. Users can browse the Featured Tree Collections or Upload Trees to your own workplace both registerially and anonymously.
Citation
Ziqi Deng, Jorge Botas, Carlos P Cantalapiedra, Ana Hernández-Plaza, Jordi Burguet-Castell and Jaime Huerta-Cepas
Nucleic Acids Research. 2022
Tree management
You can upload one or more tree at once, with or without their multiple sequence alignments. Many trees can be grouped into collections for browsing and searching options.
Edit tree
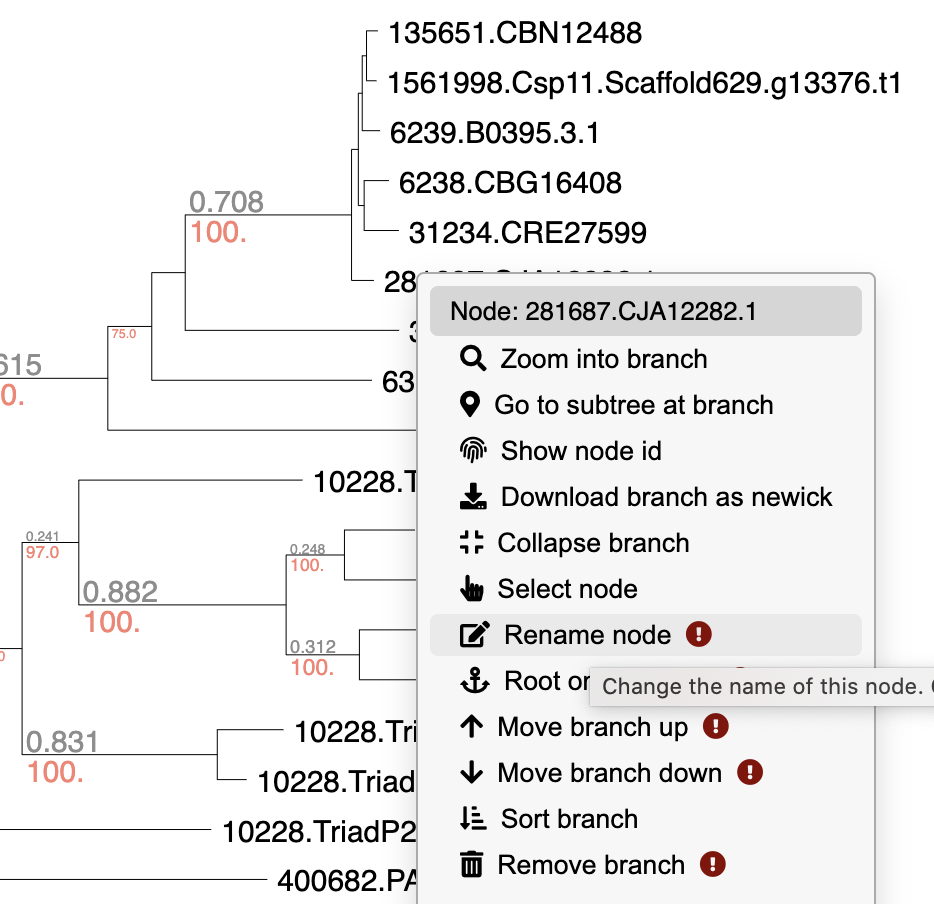
While viewing a tree, right click on any branch or node to move it, delete it, rename it, or more. You can also extract particular clades, re-root trees, or download subtrees in Newick format.
Annotate phylogenies
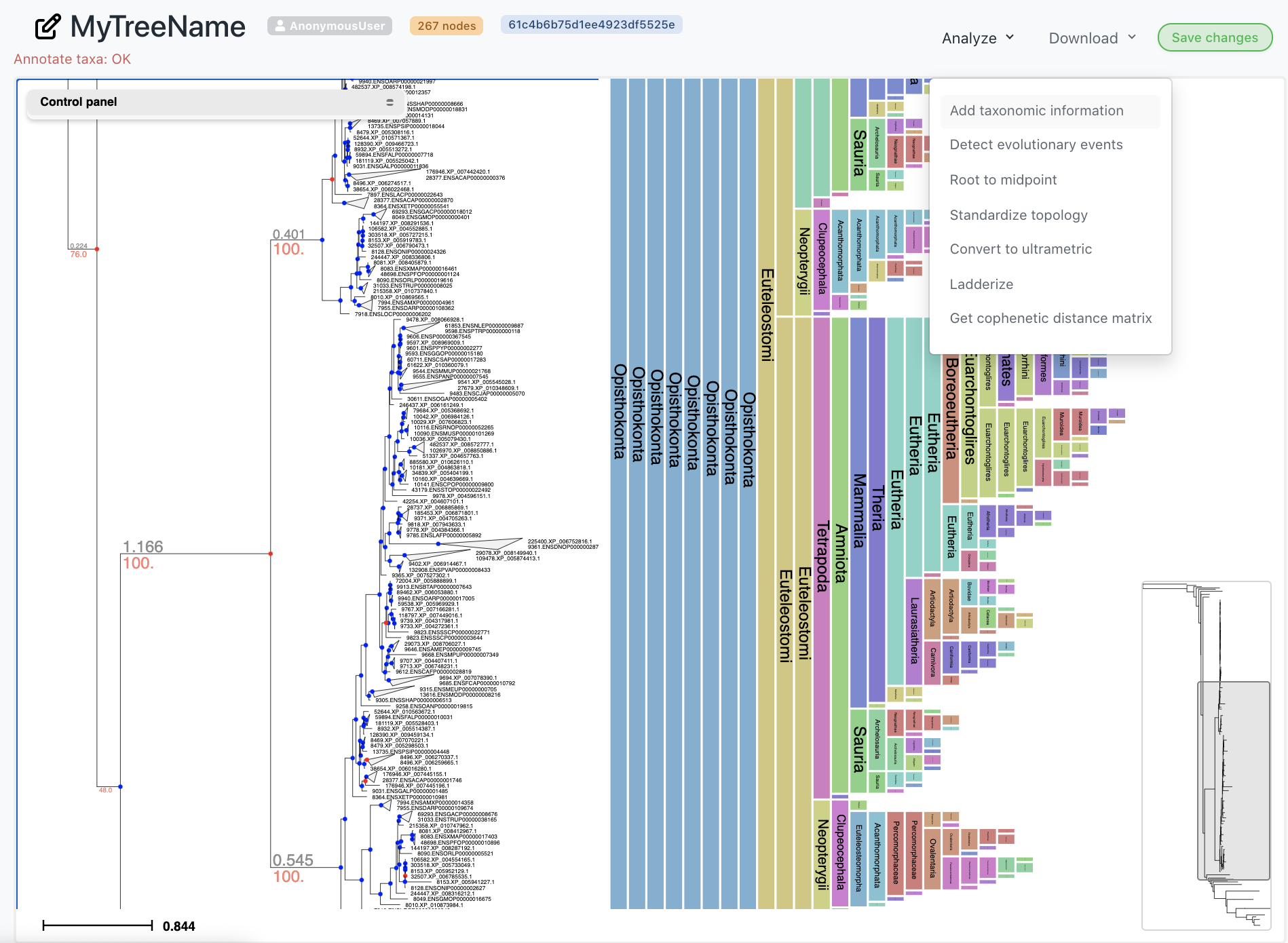
While viewing a tree, select any annotation option from the Analysis menu. You can extract species names automatically from node names and infer NCBI taxonomic information as well as speciation and duplication nodes in gene trees.
Compare Trees
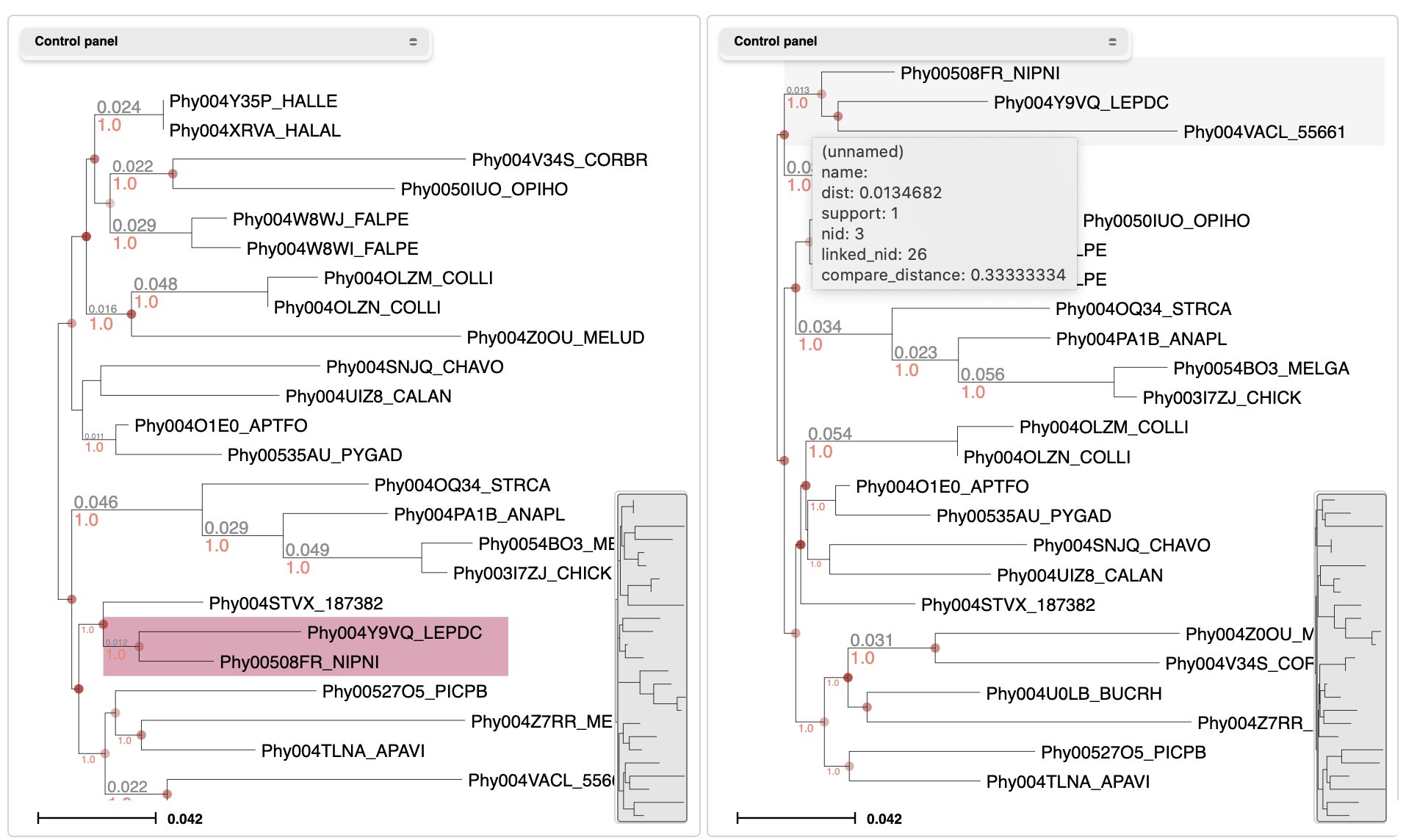
Select Tree comparison from the Tools menu in the header bar. Upload two trees with similar topologies and compare them graphically. Other metrics like the Robinson Foulds distance will be computed.
Link trees with molecular sequences
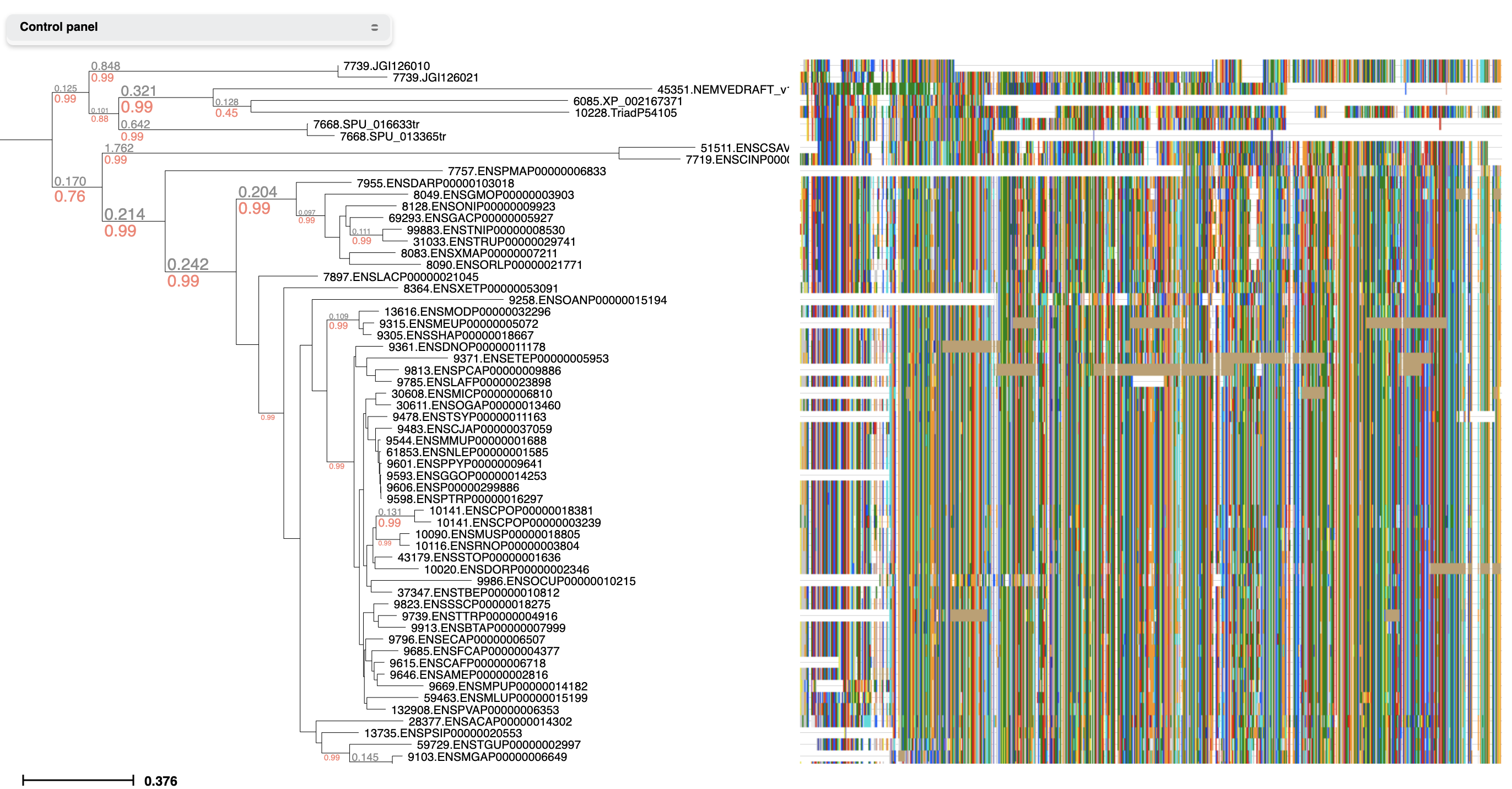
While viewing a tree, select 'Link to Alignment' from the Analysis menu. Then activate the alignment layout to explore both tree topology and alignment in an interactive manner.
Phylogenetic reconstruction
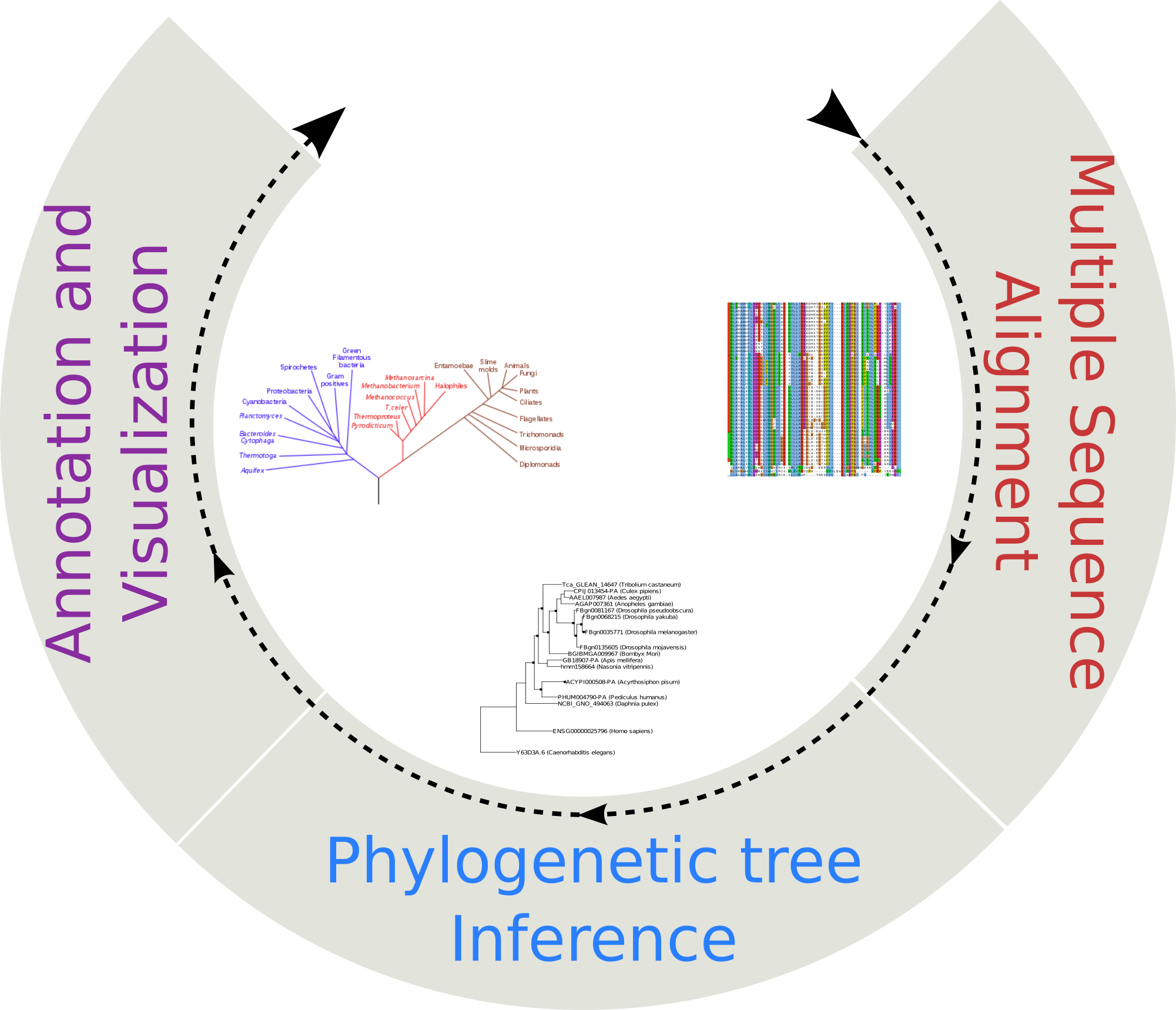
Select Phylogenetic reconstruction from the Tools menu. Submit nucleotide or amino acids sequences in FASTA format and infer a phylogeny using ETE-Toolkit predefined workflows.
Phylogenetic sequence placement
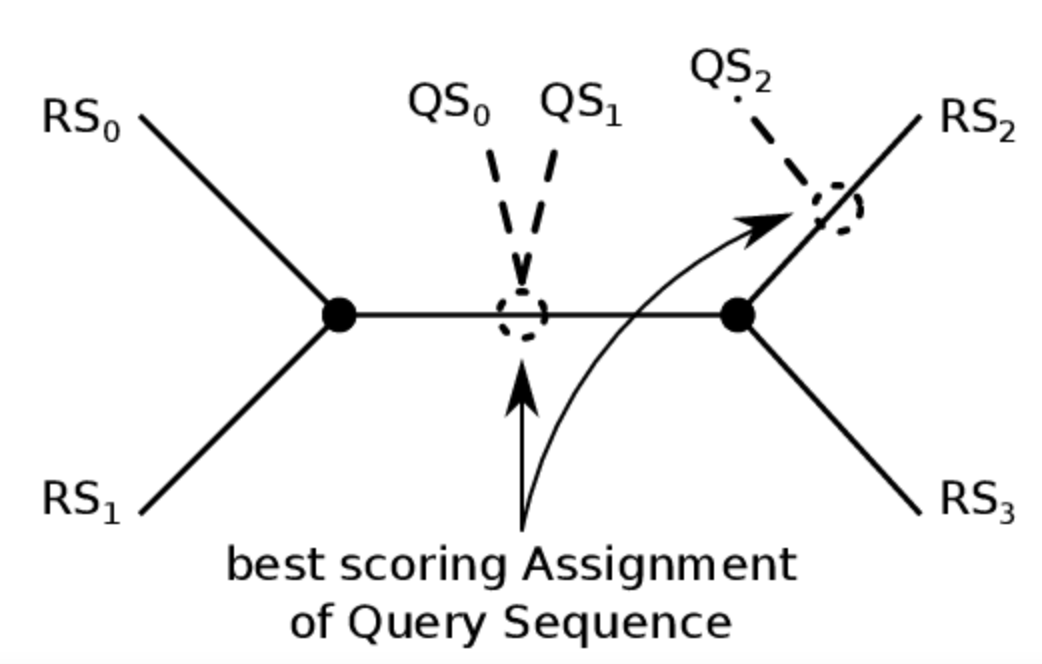
Select Sequence placement from the Tools menu. Submit a protein sequence and place it in its gene-family tree.
Query taxonomic databases
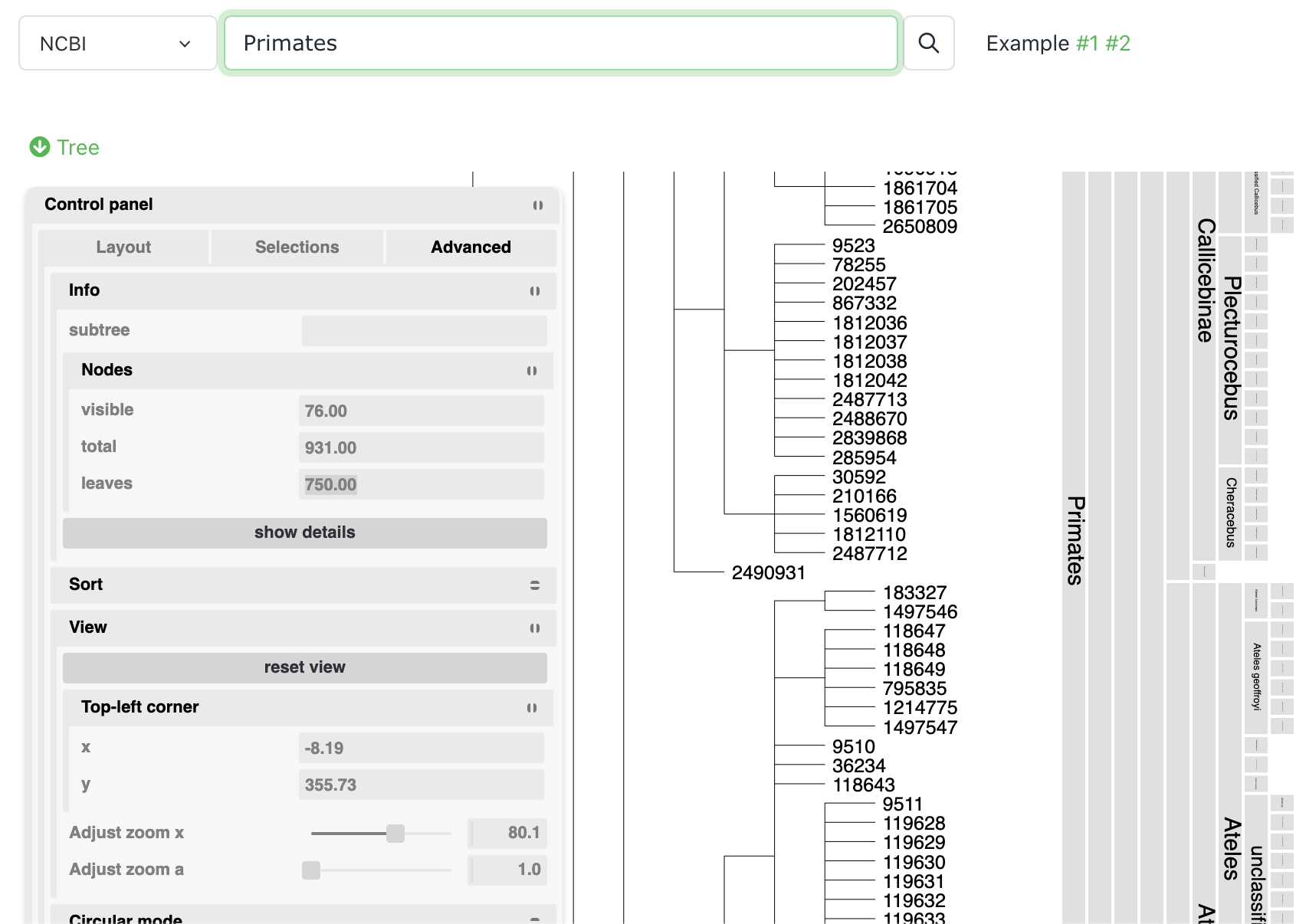
Select Taxonomy queries from the Tools menu, and query either NCBI Taxonomy tree or GTDB global phylogeny. You can then download the subtree of the queried terms.

